Building the scientific evidence base of food quality
To unlock the potential of food for supporting the health of people and the planet, we must build the scientific evidence base.
We are supporting scientific research and interventions that apply PTFI multi-omics tools on how environmental, agricultural, and cultural factors impact food quality based on diverse components and implications for human and planetary health.
Core to these efforts is our collection of rich metadata to characterize foods. We are taking a food systems approach of capturing the multitude of biophysical and sociocultural variables that impact food components. This metadata will enable scientists around the world to use PTFI data to answer big research questions on society’s pressing food system challenges including biodiversity loss, climate change, malnutrition, and unsustainable agricultural production.
Our ultimate goal is to generate knowledge on how our food system can become more diverse, resilient, inclusive, connected, participatory, and responsible.
Big Data Research Questions
We are designing the PTFI data platform to build the evidence base to answer big data questions on pressing societal issues, including:
- What is in the food we eat?
- How does food quality vary based on climate, geography, and how food is produced and processed?
- What are the implications of variation in food quality for people and the planet?
Ultimately, we seek to tap into the PTFI data to ask questions to inform evidence-based programs, guidelines, and policies such as: How can we produce food in ways to be more nourishing, desirable, and affordable while using fewer resources, conserving biodiversity, and reducing carbon emissions?
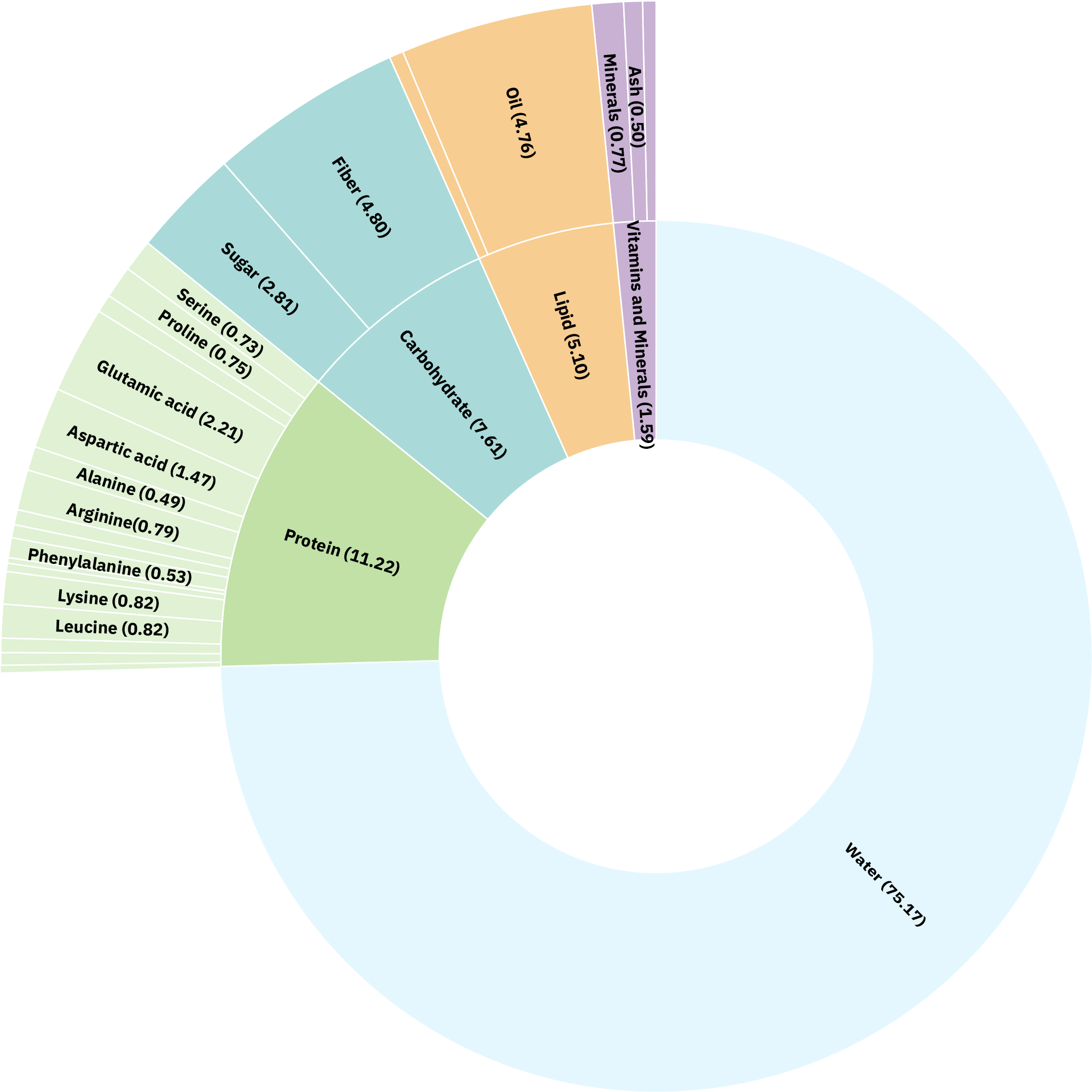
Of a total of 100%, Water is 75.17%.
Vitamins and minerals is 1.59%,
Lipid is 5.10%,
Cabohydrate is 7.61%,
Protein is 11.22%.
Of the vitamins and minerals,
Ash is 0.50%
Minerals are 0.77%
Of the Lipids,
Oil is 4.76%
Of the Carbohydrates,
Fiber is 4.80%,
Sugar is 2.81%
Of the Proteins,
Serine is 0.73%,
Proline is 0.75%,
Glutamic acid is 2.21%,
Aspartic acid is 1.47%,
Alanine is 0.49%,
Arginine is 0.79%,
Phenylalanine is 0.53%,
Lysine is 0.82%,
Leucine is 0.82%
What’s in a Plum?
It seems like a simple question, but the answer is complex! The figure to the left shows the multiple levels of detail that we are assembling for each food represented in the Periodic Table of Food database.
The inner ring shows the proportional composition of macronutrients, including protein, carbohydrates, fat, ash, and water.
The second ring lets us explore elements like the amino acid composition found within the total protein fraction of the plum, and the proportion of fiber, sugar, and starch within the carbohydrate fraction. We can dive even deeper into these data by looking at the types of fiber present, as well as the linkages of sugars to each fiber type, for example.
By layering the various types of data for each food, database users can choose the appropriate level of detail for the types of questions they want to answer. In this way, the PTFI multi-omics platforms greatly augment existing methods and the decades of data that has already been assembled across the world.
When this is combined with the metadata collected with each sample, it’s easy to imagine an atlas of deep food composition knowledge that will enable data-driven solutions for human and planetary health.
As increasing numbers of food types are analyzed using standardized tools for mapping food quality, we will increase the database on food biodiversity and exponentially expand our ability to explore both known and unknown molecules in our food.
To address our most pressing human and planetary health challenges, we need to know more about what our food is made of.
Advanced analytical methods and instrumentation are enabling the generation of deep comprehensive data on the thousands of components in food to complement existing methods for essential nutrient analysis. However, these methods were not yet standardized, so data generated across labs are not directly comparable. For instance, the PTFI sent identical food samples to multiple labs for identification of small biomolecules using metabolomics. Findings showed very little overlapping results between labs, despite strong reproducibility within each lab.
PTFI has tackled this challenge head-on by developing standardized tools for identifying the diverse components in food. We are distributing these tools to labs around the world that can then generate data to better understand the quality of our global food supply and how to improve it for the betterment of life on the planet.

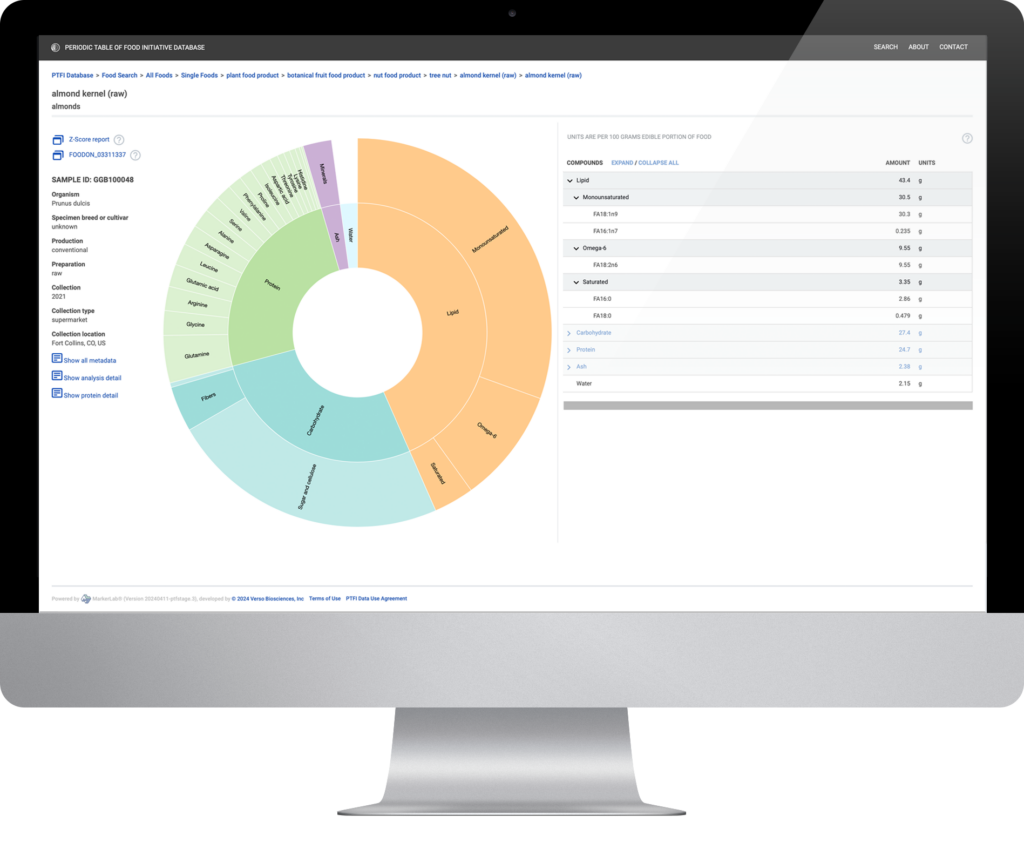
Multi-omics data organized and presented in an intuitive, engaging and enlightening format.
MarkerLab is a web-based data visualization platform that makes
multi-omics data accessible, interpretable, and valuable for labs and researchers.
Explore PTFI Data with MarkerLab
The Periodic Table of Food Initiative is mapping food quality based on multi-omics data around the globe to improve human and planetary health. Researchers can use MarkerLab to search the PTFI database and explore food biomolecular data using a variety of visualization options. In addition to interactive data plots and tables for individual samples, MarkerLab also provides the option to compare foods and compounds of interest across the database.
Visit the PTFI Research Hub and register your account to access PTFI data with MarkerLab.

The American Heart Association’s Precision Medicine Platform is a cloud-based technology solution that allows researchers to collaborate and analyze large datasets from any computer in the world using a secure environment and the power of machine learning.
Within the research interface, users can request access to assorted datasets, including the Periodic Table of Food Initiative database to accelerate findings into impactful discoveries.
PTFI DATA
Researchers can analyze raw PTFI data to gain insights and run analyses utilizing pre-installed analytic tools. The AHA Data Science team provides a tutorial notebook in the workspace to guide users on how to view the dataset and run summary statistics. This tutorial is intended to help jumpstart analysis of the PTFI data on the Precision Medicine Platform.
Visit the Precision Medicine Platform’s PTFI data resource page for more information about requesting a workspace to analyze the PTFI data.
The quality of our food is impacted by the environment in which it is cultivated – as well as how we process, store, and prepare it.
The era of big data has led to the development of metadata, a class of information to describe and contextualize other information systematically with the goals of increasing data accessibility and enabling discovery of novel patterns. Integration of complex data sets is one of the most challenging tasks for interdisciplinary research and digital data collections.
The PTFI is developing standardized protocols to collect metadata that characterize food and food systems, including ecological, socio-cultural, economic, and health attributes.
Our approach to providing highly standardized data on the components in food begins at point-of-collection, with protocols on how to collect foods, before foods are analyzed with our multi-omics tools and processed through our PTFI data pipeline.
In accordance with the FAIR data principles (findable, accessible, interoperable, and reusable), we have developed protocols to collect customized metadata to contextualize food along with following internationally recognized ontologies, typologies, and vocabulary.
Food is a biological resource, so Access and Benefit Sharing (ABS) is core to our approach of providing food data as a global public good.
ABS is an internationally recognized principle that refers to countries’ sovereign rights over their biological resources and the capacity to regulate access to the genetic resources and derived digital sequence information (DSI) found in their territories. Food data users have an obligation to share the benefits arising from these resources with the providing countries in a fair and equitable manner.
PTFI has developed compliance protocols for the use of food samples in food composition analysis that may be subject to national ABS laws that implement international ABS agreements. PTFI partners commit to exercise compliance and due diligence regarding applicable ABS laws in each of the countries where food is procured.
Managers of the PTFI database will also adopt measures to ensure that all data that is made available is published in accordance with applicable ABS laws and agreements.
PTFI’s analytical approach is characterized by its collaborative, standardized, and distributable nature.
Collaborative
PTFI takes a collaborative approach to standardizing Foodomics tools, engaging lab partners around the globe in the evaluation, development and validation of multi-omics protocols. These platforms currently include untargeted metabolomics, lipidomics, ionomics and fatty acid analysis. Additional platforms currently in development include glycomics, targeted metabolomics, proteomics and aromatics. More about our partners.
Standardized
PTFI has four key areas of standardization to enable distributable analytical methods to labs around the world, including:
- Standardized protocols for each multi-omics platform, which will be publicly accessible at launch
- Custom internal standards to accompany the PTFI methods that allow for harmonization of data across labs, allowing the global ecosystem of PTFI partners to generate and contribute food composition data to the PTFI database
- An expanding cloud-based chemical library that allows for confident annotation of features detected in foods, without the need for labs to create their own chemical libraries
- Centralized data pipeline, allowing labs to upload raw data from their instruments via the PTFI lab portal for data processing, retention time alignment and final data assembly
Distributable
PTFI’s overarching goal is to enable labs around the world to access methods, participate in food composition analysis, and contribute to building the largest database of food biomolecular composition to date.
We’re currently working with our nine Centers of Excellence around the globe to onboard PTFI methods and enable them to serve as PTFI hubs in their geographic regions. Once launched, the PTFI methods, reagents, chemical library, and data pipeline will also be available for individual labs to participate in this global effort.
The PTFI is supporting a research portfolio that applies standardized tools to map food quality in the following priority areas:
Agroecology and Regenerative Agriculture
Food composition data can provide quantitative evidence for comparing agricultural systems in ways that take nutrition and health attributes of what is produced into account.
We support several projects that examine the benefits for ecosystems and communities of agricultural systems that are grounded in principles of Indigenous Peoples’ food systems, agroecology, and regenerative agriculture.
Climate Change
Food composition data collected over time and space can serve as an indicator and predictor of the impact of climate change on food quality. It can also provide evidence on the benefits of climate change solutions for food quality. The PTFI database can serve as a centralized global resource to track the impacts of climate change and its solutions on food quality and safety.
Diet-Related Chronic Disease and Food is Medicine Solutions
Food composition data can inform the design of diets for mitigating diet-related chronic disease that are place-based and accessible. The PTFI is working to identify the biomolecular components in food that provide health attributes. We are also beginning to integrate our tools for human serum in Food is Medicine (FIM) clinical trials, to contribute to the understanding of effectiveness of implementing FIM interventions.
Our planet has over 30,000 edible species, and this diversity is critical for food security and building the resilience of ecosystems and communities. But right now, almost half of all calories consumed by humans comes from just three sources: rice, corn, and wheat– and many other edible species are disappearing before we even understand what they offer for human health and the environment.
PTFI’s Inspirational Foods project sought to draw attention to the diversity of food on our planet. We asked a group of global experts to nominate a list of inspiring foods to represent a planetary cornucopia, based on: contribution to the human diet; cultural relevance; geographic, functional, and nutritional diversity; and innovation potential.
The experts engaged with stakeholders across the globe to nominate a rich cadre of geographically- and culturally-specific foods. Surveys were distributed through key regional partners to hear from farmers, women’s groups, and Indigenous peoples.
Just 22% and 25% of these foods, respectively, are included in USDA FoodData Central and FAO’s INFOODS nutrient databases. This highlights the tremendous opportunity that exists to expand our knowledge of the composition of the world’s edible biodiversity.
FUTURE DIRECTION: 1,600+ INSPIRATIONAL FOODS
List of Foods
1,650 foods nominated by regional experts
1/3 nominated by multiple sources
Food Groups
Plants: Fruits, Vegetables, Nuts and Seeds; Beans and Pulses
Animals: Land animal products; Aquatic animal products
Unique Foods
362 reported USDA Food Data Central
405 in FAO’s nutrient databases
The key dimensions considered in developing the Inspirational Food List are as follows:
| Category | Criteria | Description |
|---|---|---|
| Biology | Where is the origin of a food in the phylogenetic tree? | Inspirational Foods include a wide spectrum of organisms across phylogenetic diversity, including bacteria, yeast, mold, algae, insects, plants, and animals. |
| Tissue | What part of organisms are used for food? | Inspirational Foods include a diversity of organism parts that are explicitly used for food including seeds, nuts, fruits, leaves, roots, eggs, muscles, etc. |
| Geography | Where do foods originate, and where do they thrive beyond their centers of origin? | Inspirational Foods cover a wide geography of native origins of foods, as well as those that established conspicuous success in other regions of the globe. |
| Consumers | Where is the origin of a food in the phylogenetic tree? | Inspirational Foods take into account the processing and use of agricultural and wild commodities, from wild to fermented and cooked. |
| Processing | How are foods treated and used after harvest? | Inspirational Foods include a wide spectrum of organisms across phylogenetic diversity, including bacteria, yeast, mold, algae, insects, plants, and animals. |
| Domestication | How has human intervention modified organisms? | Inspirational Foods capture the influence of human activity on the genetic evolution of foods, including trait-based selection. |
| Proportional Abundance | Which foods are the center of a meal and which are tiny fractions that are frequently consumed? | Inspirational Foods represent the breadth and depth within consumption patterns. |
| Affordability | Which foods are luxuries and which are staples? | Inspirational Foods include those that are widely and narrowly accessible, as a means to understand the relationship between food composition and cost and support decision-makers with data on how to best find nutrient sources to meet dietary and health needs. |
| Frequency | Which foods are consumed on a regular basis and which are associated with rare festive events, life transitions, spiritual celebrations? | Inspirational Foods include those used as dietary staples to underutilized crops and celebration foods including those that have a history of distinct value to societies beyond their nutritional value. |
| Complementarity | Where is the origin of a food in the phylogenetic tree? | Inspirational Foods include a wide spectrum of organisms across phylogenetic diversity, including bacteria, yeast, mold, algae, insects, plants, and animals. |






